Bias Correction¶
This notebook demonstrates z-score bias correction in 4 steps:
Look at zonal wind data
Filtering the data to the extent of Colorado
Aligning the datasets in time
A z-score bias correction method
Set up Workspace¶
Import all necessary modules and name the variable we will be looking at (zonal wind speed, uas).
[1]:
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
from scipy import interpolate, stats
import xesmf as xe
from datetime import datetime, timedelta
from ngallery_utils import DATASETS
%matplotlib inline
var = 'uas'
1. Look at Data¶
We have 3 datasets for this correction: a historical model, a future rcp85 model, and measurements.
Ultimately, the historical model will be compared to the measurements to predict an adjustment to the future rcp85 model to be more like future measurements.
[2]:
hist_file = DATASETS.fetch("uas.hist.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.Colorado.nc")
rcp85_file = DATASETS.fetch("uas.rcp85.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.Colorado.nc")
meas_file = DATASETS.fetch("uas.gridMET.NAM-44i.Colorado.nc")
Downloading file 'uas.hist.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.Colorado.nc' from 'ftp://ftp.cgd.ucar.edu/archive/aletheia-data/tutorial-data/uas.hist.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.Colorado.nc' to '/home/jovyan/aletheia-data/tutorial-data'.
Downloading file 'uas.rcp85.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.Colorado.nc' from 'ftp://ftp.cgd.ucar.edu/archive/aletheia-data/tutorial-data/uas.rcp85.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.Colorado.nc' to '/home/jovyan/aletheia-data/tutorial-data'.
Downloading file 'uas.gridMET.NAM-44i.Colorado.nc' from 'ftp://ftp.cgd.ucar.edu/archive/aletheia-data/tutorial-data/uas.gridMET.NAM-44i.Colorado.nc' to '/home/jovyan/aletheia-data/tutorial-data'.
[3]:
ds_hist = xr.open_dataset(hist_file)
ds_hist
[3]:
<xarray.Dataset>
Dimensions: (bnds: 2, lat: 8, lon: 16, time: 20440)
Coordinates:
* time (time) object 1950-01-01 12:00:00 ... 2005-12-31 12:00:00
* lat (lat) float64 37.25 37.75 38.25 38.75 39.25 39.75 40.25 40.75
* lon (lon) float64 -109.8 -109.2 -108.8 ... -103.2 -102.8 -102.2
Dimensions without coordinates: bnds
Data variables:
uas (time, lat, lon) float32 ...
time_bnds (time, bnds) object 1950-01-01 00:00:00 ... 2006-01-01 00:00:00
Attributes:
Conventions: CF-1.4
institution: Universite du Quebec a Montreal
contact: Winger.Katja@uqam.ca
comment: CORDEX North America CRCM5 v333 0.44 deg ...
model: CRCM5 (dynamics GEM v_3.3.3, physics RPN ...
model_grid: rotated lat-lon 236x241 incl. 10p pilot a...
geophysical_fields: orography: USGS / land use cover: USGS / ...
physics: land: CLASS3.5+, 26L, bottom at 60m / lak...
forcing: GHG: CO2,CH4,N2O,CFC11,effective CFC12
creation_date: 2012-09-06
experiment: historical
experiment_id: historical
driving_model_ensemble_member: r1i1p1
driving_experiment_name: historical
frequency: day
institute_id: UQAM
rcm_version_id: v1
project_id: CORDEX
CORDEX_domain: NAM-44
product: output
references: http://www.mrcc.uqam.ca
history: Fri Mar 29 14:03:34 2019: ncrcat -O -o /g...
NCO: netCDF Operators version 4.7.4 (http://nc...
model_id: UQAM-CRCM5
driving_experiment: CCCma-CanESM2,historical,r1i1p1
driving_model_id: CCCma-CanESM2
history_of_appended_files: Wed Sep 5 00:03:57 2018: Appended file t...
tracking_id: 5718ce6b-2ffb-4efb-9b49-f123e4184106
nco_openmp_thread_number: 1
id: doi:10.5065/D6SJ1JCH
title: NA-CORDEX Raw NAM-44i CRCM5-UQAM CanESM2 ...
version: 1.0- bnds: 2
- lat: 8
- lon: 16
- time: 20440
- time(time)object1950-01-01 12:00:00 ... 2005-12-...
- actual_range :
- [ 31.5 395.5]
- coordinate_defines :
- point
- bounds :
- time_bnds
- delta_t :
- 0000-00-01 00:00:00
- axis :
- T
- standard_name :
- time
- long_name :
- time
array([cftime.DatetimeNoLeap(1950, 1, 1, 12, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 2, 12, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 3, 12, 0, 0, 0), ..., cftime.DatetimeNoLeap(2005, 12, 29, 12, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 30, 12, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 31, 12, 0, 0, 0)], dtype=object) - lat(lat)float6437.25 37.75 38.25 ... 40.25 40.75
- long_name :
- latitude
- units :
- degrees_north
- standard_name :
- latitude
array([37.25, 37.75, 38.25, 38.75, 39.25, 39.75, 40.25, 40.75])
- lon(lon)float64-109.8 -109.2 ... -102.8 -102.2
- long_name :
- longitude
- units :
- degrees_east
- standard_name :
- longitude
array([-109.75, -109.25, -108.75, -108.25, -107.75, -107.25, -106.75, -106.25, -105.75, -105.25, -104.75, -104.25, -103.75, -103.25, -102.75, -102.25])
- uas(time, lat, lon)float32...
- units :
- m s-1
- standard_name :
- eastward_wind
- actual_range :
- [-28.95829057 28.48435832]
- grid_desc :
- rotated_pole
- long_name :
- Eastward Near-Surface Wind
- cell_methods :
- time: mean
- remap :
- remapped via ESMF_regrid_with_weights: Higher-order Patch
[2616320 values with dtype=float32]
- time_bnds(time, bnds)object...
array([[cftime.DatetimeNoLeap(1950, 1, 1, 0, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 2, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(1950, 1, 2, 0, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 3, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(1950, 1, 3, 0, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 4, 0, 0, 0, 0)], ..., [cftime.DatetimeNoLeap(2005, 12, 29, 0, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 30, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(2005, 12, 30, 0, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 31, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(2005, 12, 31, 0, 0, 0, 0), cftime.DatetimeNoLeap(2006, 1, 1, 0, 0, 0, 0)]], dtype=object)
- Conventions :
- CF-1.4
- institution :
- Universite du Quebec a Montreal
- contact :
- Winger.Katja@uqam.ca
- comment :
- CORDEX North America CRCM5 v333 0.44 deg CCCMA-CanESM2 L56
- model :
- CRCM5 (dynamics GEM v_3.3.3, physics RPN 5.0.4)
- model_grid :
- rotated lat-lon 236x241 incl. 10p pilot and 10p blending
- geophysical_fields :
- orography: USGS / land use cover: USGS / sand & clay: ECOCLIMAP
- physics :
- land: CLASS3.5+, 26L, bottom at 60m / lakes: FLake / precipitation: bourge3d / vertical diffusion: clef / shallow convection : conres & ktrsnt / deep convection: Kain-Fritsch / radiation : cccmarad
- forcing :
- GHG: CO2,CH4,N2O,CFC11,effective CFC12
- creation_date :
- 2012-09-06
- experiment :
- historical
- experiment_id :
- historical
- driving_model_ensemble_member :
- r1i1p1
- driving_experiment_name :
- historical
- frequency :
- day
- institute_id :
- UQAM
- rcm_version_id :
- v1
- project_id :
- CORDEX
- CORDEX_domain :
- NAM-44
- product :
- output
- references :
- http://www.mrcc.uqam.ca
- history :
- Fri Mar 29 14:03:34 2019: ncrcat -O -o /glade/collections/cdg/data/cordex/data2/raw/NAM-44i/day/CRCM5-UQAM/CanESM2/hist/uas/uas.hist.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.nc -p /glade/collections/cdg/work/cordex/esgf/crcm5-uqam/canesm2/nam-44i/hist/day uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19510101-19551231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19560101-19601231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19610101-19651231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19660101-19701231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19710101-19751231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19760101-19801231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19810101-19851231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19860101-19901231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19910101-19951231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19960101-20001231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_20010101-20051231.nc Wed Sep 5 00:03:57 2018: ncks -A time_bnds.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc Wed Sep 5 00:03:57 2018: ncks -v time_bnds ../uas_NAM-44_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc time_bnds.nc Thu Sep 6 22:52:43 2012: ncks -a -x -v plev,lev -4 -L 1 uas_NAM-44_CCCMA-CanESM2_historical_r1i1p1_UQAM_CRCM_v1_day_19500101-19501231.nc /home/winger/work/CORDEX/NorthAmerica_0.44deg_CanHisto_B1/NetCDF/trav_1736/tmp_1736
- NCO :
- netCDF Operators version 4.7.4 (http://nco.sf.net)
- model_id :
- UQAM-CRCM5
- driving_experiment :
- CCCma-CanESM2,historical,r1i1p1
- driving_model_id :
- CCCma-CanESM2
- history_of_appended_files :
- Wed Sep 5 00:03:57 2018: Appended file time_bnds.nc had following "history" attribute: Wed Sep 5 00:03:57 2018: ncks -v time_bnds ../uas_NAM-44_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc time_bnds.nc Thu Sep 6 22:52:43 2012: ncks -a -x -v plev,lev -4 -L 1 uas_NAM-44_CCCMA-CanESM2_historical_r1i1p1_UQAM_CRCM_v1_day_19500101-19501231.nc /home/winger/work/CORDEX/NorthAmerica_0.44deg_CanHisto_B1/NetCDF/trav_1736/tmp_1736
- tracking_id :
- 5718ce6b-2ffb-4efb-9b49-f123e4184106
- nco_openmp_thread_number :
- 1
- id :
- doi:10.5065/D6SJ1JCH
- title :
- NA-CORDEX Raw NAM-44i CRCM5-UQAM CanESM2 Hist Daily Mean 10m Eastward Wind Velocity
- version :
- 1.0
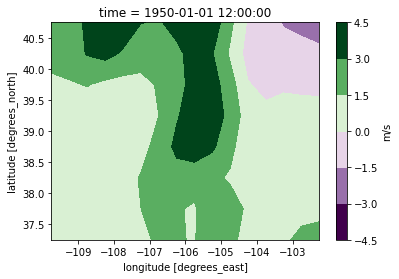
[4]:
ds_hist[var].isel(time=0).plot.contourf(x="lon", y="lat", cmap="PRGn", cbar_kwargs={"label": "m/s"})
[4]:
<matplotlib.contour.QuadContourSet at 0x7fc0757430a0>
[5]:
ds_rcp85 = xr.open_dataset(rcp85_file)
ds_rcp85
[5]:
<xarray.Dataset>
Dimensions: (bnds: 2, lat: 8, lon: 16, time: 20440)
Coordinates:
* time (time) object 1950-01-01 12:00:00 ... 2005-12-31 12:00:00
* lat (lat) float64 37.25 37.75 38.25 38.75 39.25 39.75 40.25 40.75
* lon (lon) float64 -109.8 -109.2 -108.8 ... -103.2 -102.8 -102.2
Dimensions without coordinates: bnds
Data variables:
uas (time, lat, lon) float32 ...
time_bnds (time, bnds) object 1950-01-01 00:00:00 ... 2006-01-01 00:00:00
Attributes:
Conventions: CF-1.4
institution: Universite du Quebec a Montreal
contact: Winger.Katja@uqam.ca
comment: CORDEX North America CRCM5 v333 0.44 deg ...
model: CRCM5 (dynamics GEM v_3.3.3, physics RPN ...
model_grid: rotated lat-lon 236x241 incl. 10p pilot a...
geophysical_fields: orography: USGS / land use cover: USGS / ...
physics: land: CLASS3.5+, 26L, bottom at 60m / lak...
forcing: GHG: CO2,CH4,N2O,CFC11,effective CFC12
creation_date: 2012-09-06
experiment: historical
experiment_id: historical
driving_model_ensemble_member: r1i1p1
driving_experiment_name: historical
frequency: day
institute_id: UQAM
rcm_version_id: v1
project_id: CORDEX
CORDEX_domain: NAM-44
product: output
references: http://www.mrcc.uqam.ca
history: Fri Mar 29 14:03:34 2019: ncrcat -O -o /g...
NCO: netCDF Operators version 4.7.4 (http://nc...
model_id: UQAM-CRCM5
driving_experiment: CCCma-CanESM2,historical,r1i1p1
driving_model_id: CCCma-CanESM2
history_of_appended_files: Wed Sep 5 00:03:57 2018: Appended file t...
tracking_id: 5718ce6b-2ffb-4efb-9b49-f123e4184106
nco_openmp_thread_number: 1
id: doi:10.5065/D6SJ1JCH
title: NA-CORDEX Raw NAM-44i CRCM5-UQAM CanESM2 ...
version: 1.0- bnds: 2
- lat: 8
- lon: 16
- time: 20440
- time(time)object1950-01-01 12:00:00 ... 2005-12-...
- actual_range :
- [ 31.5 395.5]
- coordinate_defines :
- point
- bounds :
- time_bnds
- delta_t :
- 0000-00-01 00:00:00
- axis :
- T
- standard_name :
- time
- long_name :
- time
array([cftime.DatetimeNoLeap(1950, 1, 1, 12, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 2, 12, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 3, 12, 0, 0, 0), ..., cftime.DatetimeNoLeap(2005, 12, 29, 12, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 30, 12, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 31, 12, 0, 0, 0)], dtype=object) - lat(lat)float6437.25 37.75 38.25 ... 40.25 40.75
- long_name :
- latitude
- units :
- degrees_north
- standard_name :
- latitude
array([37.25, 37.75, 38.25, 38.75, 39.25, 39.75, 40.25, 40.75])
- lon(lon)float64-109.8 -109.2 ... -102.8 -102.2
- long_name :
- longitude
- units :
- degrees_east
- standard_name :
- longitude
array([-109.75, -109.25, -108.75, -108.25, -107.75, -107.25, -106.75, -106.25, -105.75, -105.25, -104.75, -104.25, -103.75, -103.25, -102.75, -102.25])
- uas(time, lat, lon)float32...
- units :
- m s-1
- standard_name :
- eastward_wind
- actual_range :
- [-28.95829057 28.48435832]
- grid_desc :
- rotated_pole
- long_name :
- Eastward Near-Surface Wind
- cell_methods :
- time: mean
- remap :
- remapped via ESMF_regrid_with_weights: Higher-order Patch
[2616320 values with dtype=float32]
- time_bnds(time, bnds)object...
array([[cftime.DatetimeNoLeap(1950, 1, 1, 0, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 2, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(1950, 1, 2, 0, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 3, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(1950, 1, 3, 0, 0, 0, 0), cftime.DatetimeNoLeap(1950, 1, 4, 0, 0, 0, 0)], ..., [cftime.DatetimeNoLeap(2005, 12, 29, 0, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 30, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(2005, 12, 30, 0, 0, 0, 0), cftime.DatetimeNoLeap(2005, 12, 31, 0, 0, 0, 0)], [cftime.DatetimeNoLeap(2005, 12, 31, 0, 0, 0, 0), cftime.DatetimeNoLeap(2006, 1, 1, 0, 0, 0, 0)]], dtype=object)
- Conventions :
- CF-1.4
- institution :
- Universite du Quebec a Montreal
- contact :
- Winger.Katja@uqam.ca
- comment :
- CORDEX North America CRCM5 v333 0.44 deg CCCMA-CanESM2 L56
- model :
- CRCM5 (dynamics GEM v_3.3.3, physics RPN 5.0.4)
- model_grid :
- rotated lat-lon 236x241 incl. 10p pilot and 10p blending
- geophysical_fields :
- orography: USGS / land use cover: USGS / sand & clay: ECOCLIMAP
- physics :
- land: CLASS3.5+, 26L, bottom at 60m / lakes: FLake / precipitation: bourge3d / vertical diffusion: clef / shallow convection : conres & ktrsnt / deep convection: Kain-Fritsch / radiation : cccmarad
- forcing :
- GHG: CO2,CH4,N2O,CFC11,effective CFC12
- creation_date :
- 2012-09-06
- experiment :
- historical
- experiment_id :
- historical
- driving_model_ensemble_member :
- r1i1p1
- driving_experiment_name :
- historical
- frequency :
- day
- institute_id :
- UQAM
- rcm_version_id :
- v1
- project_id :
- CORDEX
- CORDEX_domain :
- NAM-44
- product :
- output
- references :
- http://www.mrcc.uqam.ca
- history :
- Fri Mar 29 14:03:34 2019: ncrcat -O -o /glade/collections/cdg/data/cordex/data2/raw/NAM-44i/day/CRCM5-UQAM/CanESM2/hist/uas/uas.hist.CanESM2.CRCM5-UQAM.day.NAM-44i.raw.nc -p /glade/collections/cdg/work/cordex/esgf/crcm5-uqam/canesm2/nam-44i/hist/day uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19510101-19551231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19560101-19601231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19610101-19651231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19660101-19701231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19710101-19751231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19760101-19801231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19810101-19851231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19860101-19901231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19910101-19951231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19960101-20001231.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_20010101-20051231.nc Wed Sep 5 00:03:57 2018: ncks -A time_bnds.nc uas_NAM-44i_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc Wed Sep 5 00:03:57 2018: ncks -v time_bnds ../uas_NAM-44_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc time_bnds.nc Thu Sep 6 22:52:43 2012: ncks -a -x -v plev,lev -4 -L 1 uas_NAM-44_CCCMA-CanESM2_historical_r1i1p1_UQAM_CRCM_v1_day_19500101-19501231.nc /home/winger/work/CORDEX/NorthAmerica_0.44deg_CanHisto_B1/NetCDF/trav_1736/tmp_1736
- NCO :
- netCDF Operators version 4.7.4 (http://nco.sf.net)
- model_id :
- UQAM-CRCM5
- driving_experiment :
- CCCma-CanESM2,historical,r1i1p1
- driving_model_id :
- CCCma-CanESM2
- history_of_appended_files :
- Wed Sep 5 00:03:57 2018: Appended file time_bnds.nc had following "history" attribute: Wed Sep 5 00:03:57 2018: ncks -v time_bnds ../uas_NAM-44_CCCma-CanESM2_historical_r1i1p1_UQAM-CRCM5_v1_day_19500101-19501231.nc time_bnds.nc Thu Sep 6 22:52:43 2012: ncks -a -x -v plev,lev -4 -L 1 uas_NAM-44_CCCMA-CanESM2_historical_r1i1p1_UQAM_CRCM_v1_day_19500101-19501231.nc /home/winger/work/CORDEX/NorthAmerica_0.44deg_CanHisto_B1/NetCDF/trav_1736/tmp_1736
- tracking_id :
- 5718ce6b-2ffb-4efb-9b49-f123e4184106
- nco_openmp_thread_number :
- 1
- id :
- doi:10.5065/D6SJ1JCH
- title :
- NA-CORDEX Raw NAM-44i CRCM5-UQAM CanESM2 Hist Daily Mean 10m Eastward Wind Velocity
- version :
- 1.0
[6]:
ds_rcp85[var].isel(time=0).plot.contourf(x="lon", y="lat", cmap="PRGn", cbar_kwargs={"label": "m/s"})
[6]:
<matplotlib.contour.QuadContourSet at 0x7fc074bb0b20>
[7]:
ds_meas = xr.open_dataset(meas_file)
ds_meas
[7]:
<xarray.Dataset>
Dimensions: (bnds: 2, lat: 8, lon: 16, time: 13880)
Coordinates:
* time (time) datetime64[ns] 1979-01-01T19:00:00 ... 2016-12-31T19:...
* lon (lon) float64 -109.8 -109.2 -108.8 ... -103.2 -102.8 -102.2
* lat (lat) float64 37.25 37.75 38.25 38.75 39.25 39.75 40.25 40.75
Dimensions without coordinates: bnds
Data variables:
uas (time, lat, lon) float32 ...
time_bnds (time, bnds) datetime64[ns] 1979-01-01T07:00:00 ... 2017-01-...
data_status (time) int8 2 2 2 2 2 2 2 2 2 2 2 2 ... 0 0 0 0 0 0 0 0 0 0 0 0
lon_bnds (lon, bnds) float32 -110.0 -109.5 -109.5 ... -102.5 -102.0
lat_bnds (lat, bnds) float32 37.0 37.5 37.5 38.0 ... 40.0 40.5 40.5 41.0
crs int32 1
Attributes:
author: John Abatzoglou - University of Idaho, jabatzo...
creation_date: 10 March 2016
Conventions: CF-1.6
title: gridMET gridded surface meteorological data
institution: University of Idaho
contact: John Abatzoglou, jabatzoglou@uidaho.edu
URL: http://www.climatologylab.org/gridmet.html
references: Abatzoglou, J.T., 2012, Development of gridded...
source: gridMET combines data from PRISM and NLDAS-2 a...
acknowledgements: CF compliance by Seth McGinnis, mcginnis@ucar.edu
uuid: 82c7e638-7fb7-4a0d-bcef-c2d18a69f8fa
history: Fri Aug 30 15:00:15 2019: ncrcat -n 38,4,1 uas...
NCO: netCDF Operators version 4.7.4 (http://nco.sf....
tracking_id: da9158a7-73d3-4e74-aeaf-c0f7d0433508
nco_openmp_thread_number: 1- bnds: 2
- lat: 8
- lon: 16
- time: 13880
- time(time)datetime64[ns]1979-01-01T19:00:00 ... 2016-12-...
- note :
- Days start/end at approximately midnight Mountain Standard Time
- standard_name :
- time
- long_name :
- time
- bounds :
- time_bnds
- axis :
- T
array(['1979-01-01T19:00:00.000000000', '1979-01-02T19:00:00.000000000', '1979-01-03T19:00:00.000000000', ..., '2016-12-29T19:00:00.000000000', '2016-12-30T19:00:00.000000000', '2016-12-31T19:00:00.000000000'], dtype='datetime64[ns]') - lon(lon)float64-109.8 -109.2 ... -102.8 -102.2
- units :
- degrees_east
- long_name :
- longitude
- standard_name :
- longitude
- bounds :
- lon_bnds
array([-109.75, -109.25, -108.75, -108.25, -107.75, -107.25, -106.75, -106.25, -105.75, -105.25, -104.75, -104.25, -103.75, -103.25, -102.75, -102.25]) - lat(lat)float6437.25 37.75 38.25 ... 40.25 40.75
- units :
- degrees_north
- long_name :
- latitude
- standard_name :
- latitude
- bounds :
- lat_bnds
array([37.25, 37.75, 38.25, 38.75, 39.25, 39.75, 40.25, 40.75])
- uas(time, lat, lon)float32...
- missing_threshold :
- 50%
- cell_methods :
- time: mean(interval: 24 hours)
- grid_mapping :
- crs
- height :
- 10 m
- units :
- m/s
- standard_name :
- eastward_wind
- long_name :
- Eastward Surface Wind Speed
[1776640 values with dtype=float32]
- time_bnds(time, bnds)datetime64[ns]...
array([['1979-01-01T07:00:00.000000000', '1979-01-02T07:00:00.000000000'], ['1979-01-02T07:00:00.000000000', '1979-01-03T07:00:00.000000000'], ['1979-01-03T07:00:00.000000000', '1979-01-04T07:00:00.000000000'], ..., ['2016-12-29T07:00:00.000000000', '2016-12-30T07:00:00.000000000'], ['2016-12-30T07:00:00.000000000', '2016-12-31T07:00:00.000000000'], ['2016-12-31T07:00:00.000000000', '2017-01-01T07:00:00.000000000']], dtype='datetime64[ns]') - data_status(time)int8...
- flag_meanings :
- permanent provisional early
- flag_values :
- [0 1 2]
array([2, 2, 2, ..., 0, 0, 0], dtype=int8)
- lon_bnds(lon, bnds)float32...
array([[-110. , -109.5], [-109.5, -109. ], [-109. , -108.5], [-108.5, -108. ], [-108. , -107.5], [-107.5, -107. ], [-107. , -106.5], [-106.5, -106. ], [-106. , -105.5], [-105.5, -105. ], [-105. , -104.5], [-104.5, -104. ], [-104. , -103.5], [-103.5, -103. ], [-103. , -102.5], [-102.5, -102. ]], dtype=float32) - lat_bnds(lat, bnds)float32...
array([[37. , 37.5], [37.5, 38. ], [38. , 38.5], [38.5, 39. ], [39. , 39.5], [39.5, 40. ], [40. , 40.5], [40.5, 41. ]], dtype=float32) - crs()int32...
- grid_mapping_name :
- latitude_longitude
- longitude_of_prime_meridian :
- 0.0
- semi_major_axis :
- 6378137.0
- inverse_flattening :
- 298.257223563
- crs_wkt :
- GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]]
- long_name :
- WGS 84
array(1, dtype=int32)
- author :
- John Abatzoglou - University of Idaho, jabatzoglou@uidaho.edu
- creation_date :
- 10 March 2016
- Conventions :
- CF-1.6
- title :
- gridMET gridded surface meteorological data
- institution :
- University of Idaho
- contact :
- John Abatzoglou, jabatzoglou@uidaho.edu
- URL :
- http://www.climatologylab.org/gridmet.html
- references :
- Abatzoglou, J.T., 2012, Development of gridded surface meteorological data for ecological applications and modeling, International Journal of Climatology, DOI: 10.1002/joc.3413
- source :
- gridMET combines data from PRISM and NLDAS-2 according to the method of Abatzoglou 2012; PRISM v.LT71m: PRISM Climate Group, Oregon State University, http://prism.oregonstate.edu; NLDAS-2 v.s4pa: NASA ESD, Goddard Earth Sciences, http://ldas.gsfc.nasa.gov/nldas/
- acknowledgements :
- CF compliance by Seth McGinnis, mcginnis@ucar.edu
- uuid :
- 82c7e638-7fb7-4a0d-bcef-c2d18a69f8fa
- history :
- Fri Aug 30 15:00:15 2019: ncrcat -n 38,4,1 uas.44i.1979.nc common/uas.gridMET.NAM-44i.nc Thu Aug 29 19:19:35 MDT 2019: ncl coarsen.ncl datafile=uas.1979.nc gridfile=grid/grid.NAM-44i.nc outfile=regrid/uas.44i.1979.nc Wed Aug 28 19:26:43 2019: ncatted -a long_name,uas,o,c,Eastward Surface Wind Speed uas.1979.nc Wed Aug 28 19:26:43 2019: ncatted -a standard_name,uas,o,c,eastward_wind uas.1979.nc Wed Aug 28 19:24:26 2019: ncks -x -v spd,dir,vas wind.1979.nc uas.1979.nc Wed Aug 28 19:23:08 2019: ncap2 -A -s vas=spd*sin((270-dir)*atan(1)/45); wind.1979.nc wind.1979.nc Wed Aug 28 19:21:48 2019: ncap2 -A -s uas=spd*cos((270-dir)*atan(1)/45); wind.1979.nc wind.1979.nc Wed Aug 28 19:21:19 2019: ncks -A dir.1979.nc wind.1979.nc Wed Aug 28 18:42:26 MDT 2019: updated for CF-compliance
- NCO :
- netCDF Operators version 4.7.4 (http://nco.sf.net)
- tracking_id :
- da9158a7-73d3-4e74-aeaf-c0f7d0433508
- nco_openmp_thread_number :
- 1
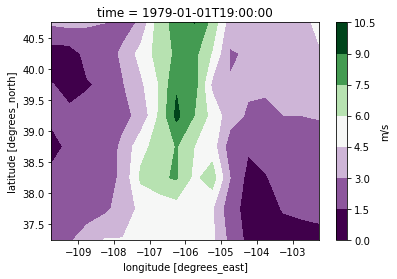
[8]:
ds_meas[var].isel(time=0).plot.contourf(x="lon", y="lat", cmap="PRGn", cbar_kwargs={"label": "m/s"})
[8]:
<matplotlib.contour.QuadContourSet at 0x7fc074e57520>
2. Filter Data¶
We limit the extent of our data to the state of Colorado simply to speed up our calculations in this example.
[9]:
wesn = [-110, -108, 39.5, 41.5]
ds_meas_flt = ds_meas.sel(lon = slice(wesn[0], wesn[1]), lat = slice(wesn[2], wesn[3]))
ds_hist_flt = ds_hist.sel(lon = slice(wesn[0], wesn[1]), lat = slice(wesn[2], wesn[3]))
ds_rcp85_flt = ds_rcp85.sel(lon = slice(wesn[0], wesn[1]), lat = slice(wesn[2], wesn[3]))
3. Align Time¶
Our two modeled datasets do not have leap years, where our measured dataset does.
There are several methods of dealing with leap years that are better for different types of data. For data that is continuous, such as wind speed, we can simply drop all December 31st from leap years (any time there is a 366th day to the year we drop day #366). This method would not be ideal for discontinous data such as precipitation.
[10]:
ds_meas_noleap = ds_meas_flt.sel(time=~((ds_meas_flt.time.dt.dayofyear == 366) ))
[11]:
def _cfnoleap_to_datetime(da):
datetimeindex = da.indexes['time'].to_datetimeindex()
ds = da#.to_dataset()
ds['time_dt']= ('time', datetimeindex)
ds = ds.swap_dims({'time': 'time_dt'})
assert len(da.time) == len(ds.time_dt)
return ds
ds_hist_dt = _cfnoleap_to_datetime(ds_hist_flt)
ds_rcp85_dt = _cfnoleap_to_datetime(ds_rcp85_flt)
<ipython-input-1-d460699fd441>:2: RuntimeWarning: Converting a CFTimeIndex with dates from a non-standard calendar, 'noleap', to a pandas.DatetimeIndex, which uses dates from the standard calendar. This may lead to subtle errors in operations that depend on the length of time between dates.
datetimeindex = da.indexes['time'].to_datetimeindex()
We also regroup our models so that the dates of the historical model line up with our historical measurements.
[12]:
def _regroup_models_bytime(ds_meas, ds_hist_dt, ds_rcp_dt):
t0_meas = ds_meas.time[0]
tn_meas = ds_meas.time[-1]
t0_fut = tn_meas.values + np.timedelta64(1, 'D')
ds_past = ds_hist_dt.sel(time_dt = slice(t0_meas, tn_meas))
ds_past = ds_past.swap_dims({'time_dt':'time'})
ds_fut_pt1 = ds_hist_dt.sel(time_dt = slice(t0_fut,None))
ds_fut = xr.concat([ds_fut_pt1[var], ds_rcp_dt[var]], 'time_dt')
ds_fut = ds_fut.swap_dims({'time_dt':'time'})
return ds_past, ds_fut
ds_past, ds_fut = _regroup_models_bytime(ds_meas_noleap, ds_hist_dt, ds_rcp85_dt)
4. Z-score Bias Correction¶
Z-Score bias correction is a good technique for target variables with Gaussian probability distributions, such as zonal wind speed.
In essence the technique:
- Finds the mean\[\overline{x} = \sum_{i=0}^N \frac{x_i}{N}\]
and standard deviation
\[\sigma = \sqrt{\frac{\sum_{i=0}^N |x_i - \overline{x}|^2}{N-1}}\]of target (measured) data and training (historical modeled) data.
Compares the difference between the statistical values to produce a shift
\[shift = \overline{x_{target}} - \overline{x_{training}}\]and scale parameter
\[scale = \sigma_{target} \div \sigma_{training}\]Applies these paramaters to the future model data to be corrected to get a new mean
\[\overline{x_{corrected}} = \overline{x_{future}} + shift\]and new standard deviation
\[\sigma_{corrected} = \sigma_{future} \times scale\]Calculates the corrected values
\[x_{corrected_{i}} = z_i \times \sigma_{corrected} + \overline{x_{corrected}}\]from the future model’s z-score values
\[z_i = \frac{x_i-\overline{x}}{\sigma}\]
In practice, if the wind was on average 3 m/s faster on the first of July in the models compared to the measurements, we would adjust the modeled data for all July 1sts in the future modeled dataset to be 3 m/s faster. And similarly for scaling the standard deviation
Bias correction methods other than zscore include:
scale (for wind speed, wave flux in air)
log (for precipitation)
range (relative humidity)
seth mcginnes’s kddm (for any bimodal distributions)
[13]:
def _reshape(ds, window_width):
split = lambda g: (g.rename({'time': 'day'})
.assign_coords(day=g.time.dt.dayofyear.values))
ds2 = ds.groupby('time.year').apply(split)
early_Jans = ds2.isel(day = slice(None,window_width//2))
late_Decs = ds2.isel(day = slice(-window_width//2,None))
ds3 = xr.concat([late_Decs,ds2,early_Jans],dim='day')
return ds3
def _calc_stats(ds, window_width):
ds_rsh = _reshape(ds, window_width)
ds_rolled = ds_rsh.rolling(day=window_width, center=True).construct('win_day')
n = window_width//2+1
ds_mean = ds_rolled.mean(dim=['year','win_day']).isel(day=slice(n,-n))
ds_std = ds_rolled.std(dim=['year','win_day']).isel(day=slice(n,-n))
ds_avyear = ds_rsh.mean(dim=['year','day'])
ds_zscore = ((ds_avyear - ds_mean) / ds_std)
return ds_mean, ds_std, ds_zscore
window_width=31
meas_mean, meas_std, meas_zscore = _calc_stats(ds_meas_noleap, window_width)
hist_mean, hist_std, hist_zscore = _calc_stats(ds_past, window_width)
[14]:
def _get_params(meas_mean, meas_std, past_mean, past_std):
shift = meas_mean - past_mean
scale = meas_std / past_std
return shift, scale
shift, scale = _get_params(meas_mean, meas_std, hist_mean, hist_std)
[15]:
def _calc_fut_stats(ds_fut, window_width):
ds_rolled = ds_fut.rolling(time=window_width, center=True).construct('win_day')
ds_mean = ds_rolled.mean(dim=['win_day'])
ds_std = ds_rolled.std(dim=['win_day'])
ds_avyear = ds_fut.mean(dim=['time'])
ds_zscore = ((ds_avyear - ds_mean) / ds_std)
return ds_mean, ds_std, ds_zscore
fut_mean, fut_std, fut_zscore = _calc_fut_stats(ds_fut, window_width)
[16]:
fut_mean_bc = fut_mean + shift.uas
fut_std_bc = fut_std * scale
[17]:
fut_corrected = (fut_zscore * fut_std_bc) + fut_mean_bc
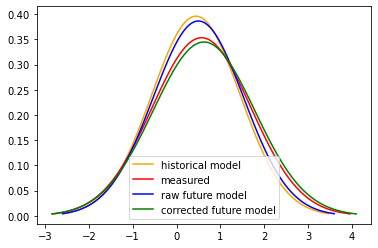
Visualize the Correction¶
[18]:
def gaus(mean, std, doy):
a = mean.sel(day=doy)
mu = a.isel(lon = 0, lat = 0)
b =std.sel(day=doy)
sigma = b.isel(lon = 0, lat = 0)
x = np.linspace(mu - 3*sigma, mu + 3*sigma, 100)
y = stats.norm.pdf(x, mu, sigma)
return x, y
[19]:
fut_typ_mean, fut_typ_std, fut_typ_zscore = _calc_stats(ds_fut, window_width)
fut_typ_mean_bc = fut_typ_mean + shift
fut_typ_std_bc = fut_typ_std * scale
doy=20
plt.figure()
x,y = gaus(hist_mean[var], hist_std[var], doy)
plt.plot(x, y, 'orange', label = 'historical model')
x,y = gaus(meas_mean[var], meas_std[var], doy)
plt.plot(x, y, 'red', label = 'measured')
x,y = gaus(fut_typ_mean, fut_typ_std, doy)
plt.plot(x, y, 'blue', label = 'raw future model')
x,y = gaus(fut_typ_mean_bc[var], fut_typ_std_bc[var], doy)
plt.plot(x, y, 'green', label = 'corrected future model')
plt.legend()
[19]:
<matplotlib.legend.Legend at 0x7fc074acd220>